Cryptosporidium
Jan Šlapeta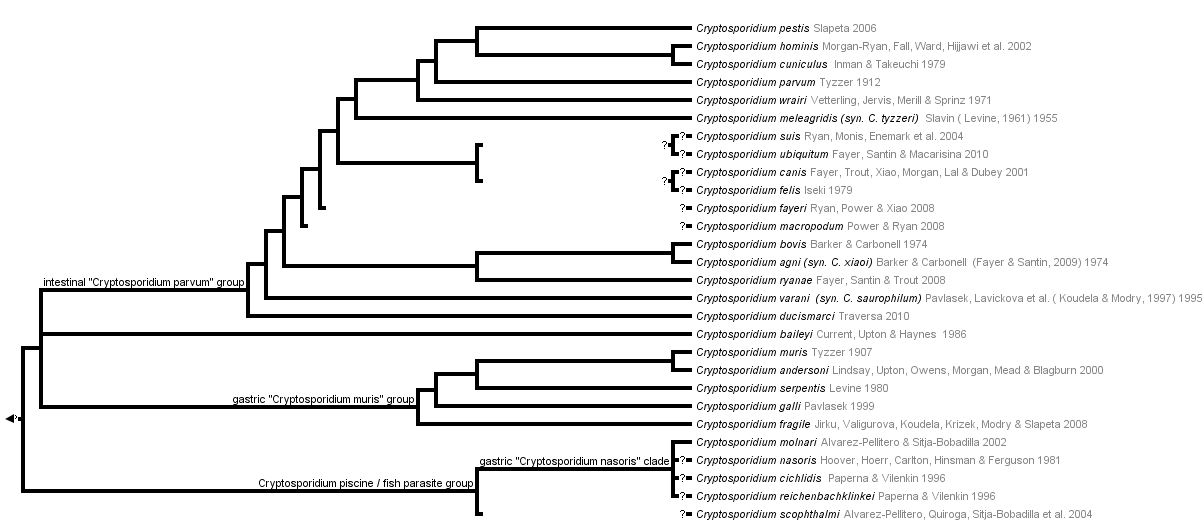

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
Family Cryptosporidiidae Leger, 1911
Genus Cryptosporidium Tyzzer, 1907
Intracellular parasites of the genus Cryptosporidium infect vertebrates, including humans, worldwide. Cryptosporidiosis is a self-limiting disease in healthy hosts but represents a life-threatening problem in immunocompromised individuals for which there is no effective treatment. In humans, the organism infects primarily the gastrointestinal epithelium producing acute watery diarrhoea. Species of Cryptosporidium parasitize specific host tissues, namely, the intestine, stomach, or trachea.
Cryptosporidium muris is the type species described from the mouse stomach. The second species differentiated from C. muris based on intestinal localisation is Cryptosporidium parvum described from mice as well.
Morphologically indistinguishable isolates are rather heterogeneous in DNA sequence, and several of those are now named as species and many acquired a genotype status. There are around 30 named species affecting mammals, birds, reptiles as well as amphibians. Plus, there are several dozens of genotypes that await formal description as species.
The species names for the major "genotypes" affecting humans formerly lumped under C. parvum are now recognised as separate species. Therefore the zoonotic genotype known in the literature as Cryptosporidium parvum "bovine genotype" is now named as the distinct species Cryptosporidium pestis. It has very low host specificity and can affect almost any vertebrate host including humans. The human specific species previously known as Cryptosporidium parvum "human genotype" is now recognised as a distinct species Cryptosporidium hominis. It is considered specific for humans transmitted by human-to-human contact. Ultimatelly, in a strict sense Cryptosporidium parvum should be restricted to rodents and is known as "mouse genotype" and has no documented capacity to infect humans.
Characteristics
Members of this genus have a direct life cycle. They develop just under the surface membrane of the host cell - this location is often called intracellular but extracytoplasmatic. Their developmental stages within the host organism are attached to the cell with a specialised attachment organelle. The life cycle consists of asexual multiplication followed by sexual development leading to development of oocysts. Oocysts develop into infective stages inside the host cell and may sustain autoinfection.
They appear to have secondarily lost the apicoplast and their mitochondrion is reduced to a minimum - the so called 'mitosome' - without any traces of DNA.
Phylogenetic Position
Cryptosporidium is traditionally considered a member of coccidia, however phylogenetic evidence shows its closer affinity with gregarines (Zhu et al. 2000, Leander et al. 2003). The whole genome sequence of Cryptosporidium species did not help to solve this dilemma, but has demonstrated Cryptosporidium's unique position within the organisms studied so far (Plasmodium, Theileria, Babesia, Eimeria, Toxoplasma, Neospora). We are still lacking comparative data from gregarines, from which this group might have evolved. Therefore, Cryptosporidium should not be classified as a coccidium. Its exact position within Apicomplexa awaits resolution. In the most recent SSU rDNA plylogenetic study, with so far the most comprehensive coverage of gregarines and phylotypes assumed to be gregarines, the position of Cryptosporidium remains unresolved (Rueckert et al. 2011). Clearly, the placement of Cryptosporidium will require a "fresh" look, i.e., new molecular markers to be obtained from gregagrines and genomes of other apicomplexan group; the most challenging part will be obtaining such markers for gregagrines (the most neglected principal group of Apicomplexa).
Discussion of Phylogenetic Relationships
Two sister groups are recognised based on molecular markes, in particular SSU rDNA, actin, HSP70 and COWP1:
- Gastric group: represented by C. muris and relatives that parasitise in the stomachs of vertebrates (other species include C. galli, C. serpentis, C. andersoni, C. fragile). Characteristically, oocysts are oval and relatively larger, ~7 um, compared to the group repreented by C. parvum.
- Intestinal group: represented by C. parvum and relatives that parasitise in the stomachs of vertebrates (other species include C. pestis, C. hominis, C. canis, C. felis, etc.). Characteristically, oocysts are round and relatively smaller, ~4-5 um, compared to the group repreented by C. muris. Moreover at the base of this group is a species afecting birds respiratoy tract - C. bailey.
A third group represented by fish Cryptosporidium species is likely to be recognised. This group represented by primarily C. molnari from stomachs of various marine and freshwater fish is a sister group to the above monophyletic gastric and intestinal groups based on SSU rDNA (Barugahare et al. 2011). Fish species are characterised by deep intramucosal develoment.
References
Barugahare, R., Becker, J. A., Landosb, M., Šlapeta, J., Dennis, M. M. 2011. Gastric cryptosporidiosis in farmed Australian Murray cod, Maccullochella peelii peelii. Aquaculture 314 (1-4), 1-6.
Carreno, R. A., D. S. Martin, and J. R. Barta. 1999. Cryptosporidium is more closely related to the gregarines than to coccidia as shown by phylogenetic analysis of apicomplexan parasites inferred using small-subunit ribosomal RNA gene sequences. Parasitology Research 85:899-904.
Leander, B. S. 2007. Molecular phylogeny and ultrastructure of Selenidium serpulae (Apicomplexa, Archigregarinia) from the calcareous tubeworm Serpula vermicularis (Annelida, Polychaeta, Sabellida). Zoologica Scripta 36:213-227.
Leander, B. S., R. Clopton, and P. Keeling. 2003. Phylogeny of gregarines (Apicomplexa) as inferred from small-subunit rDNA and beta-tubulin. International Journal of Systematic and Evolutionary Microbiology 53:345-354.
Leander, B. S., and P. J. Keeling. 2003. Morphostasis in alveolate evolution. Trends in Ecology & Evolution 18:395-402.
Rueckert, S., Simdyanov, T.G, Aleoshin, V.V., Leander, B.S. 2011. Identification of a Divergent Environmental DNA Sequence Clade Using the Phylogeny of Gregarine Parasites (Apicomplexa) from Crustacean Hosts. PLoS ONE 6(3): e18163. Šlapeta, J. 2006. Cryptosporidium species found in cattle: a proposal for a new species. Trends in Parasitology 22:469-474.
Šlapeta, J. 2009. Centenary of the genus Cryptosporidium: from morphological to molecular species identification. pp. 31-50; In: Edited by M. G. Ortega-Pierres, S. Caccio, R. Fayer, T. Mank, H. Smith, R. C. A. Thompson (Eds). Giardia and Crytosporidium, CABI Publishing.
Šlapeta, J., and J. S. Keithly. 2004. Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria. Eukaryotic Cell 3:483-494.
Valigurova, A., M. Jirku, B. Koudela, M. Gelnar, D. Modry, and J. Šlapeta. 2008. Cryptosporidia: Epicellular parasites embraced by the host cell membrane. International Journal for Parasitology 38:913-922.
Zhu, G., J. S. Keithly, and H. Philippe. 2000. What is the phylogenetic position of Cryptosporidium? International Journal of Systematic and Evolutionary Microbiology 50:1673-1681.
Title Illustrations

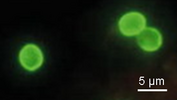
| Scientific Name | Cryptosporidium |
|---|---|
| Comments | Direct immunoflorescence antibody staining of Cryptosporidium oocysts in fecal sample. |
| Specimen Condition | Live Specimen |
| Life Cycle Stage | oocyst |
| Image Use |
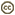 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Jan Šlapeta

|
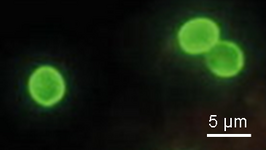
| Scientific Name | Cryptosporidium sp. in a calf intestine |
|---|---|
| Comments | Cryptosporidium developiong on the surface (intracellular but extracytoplasmatic) of the enterocysts of affected calf. Cryptosporidium appears as small round stages on the mucosal lining. |
| Specimen Condition | Dead Specimen |
| Image Use |
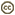 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Jan Šlapeta

|
| Scientific Name | Cryptosporidium sp. ex short-beaked echidna |
|---|---|
| Location | Sydney |
| Specimen Condition | Live Specimen |
| Image Use |
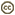 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© Jan Šlapeta

|
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Jan Šlapeta

Faculty of Veterinary Science, University of Sydney, New South Wales, Australia
Correspondence regarding this page should be directed to Jan Šlapeta at
Page copyright © 2011 Jan Šlapeta
 Page: Tree of Life
Cryptosporidium .
Authored by
Jan Šlapeta.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Cryptosporidium .
Authored by
Jan Šlapeta.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 18 May 2011
- Content changed 22 May 2011
Citing this page:
Šlapeta, Jan. 2011. Cryptosporidium . Version 22 May 2011 (under construction). http://tolweb.org/Cryptosporidium/124803/2011.05.22 in The Tree of Life Web Project, http://tolweb.org/







 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site